The Center for Genomics Sciences Imaging Facility
|
|

The new imaging facility consists of a state of the art Leica TCS SP2 AOBS filter free spectral confocal system with both inverted and upright microscopy capabilities. The system is equipped with tissue culturing apparatus for the study of mucosal biofilms.

The planned purchase of the Leica Fixed Stage Automated (LFSA) microscope will facilitate the observation of biofilm infections in whole animals.
|
The new confocal imaging facility at the Center for Genomics Sciences (CGS) is dedicated primarily to the study of bacterial biofilms and human disease. The facility combines confocal scanning laser microscopy and real-time time lapse imaging of bacterial biofilms growing in situ without having to fix or dehydrate the sample significantly reducing artifacts. The new CGS Imaging Facility will improve our ability to investigate dynamic, living biofilms in situ in three dimensions and in real time.
Imaging is crucial to the investigation of biofilms. Such studies lead to a better understanding of the complex nature of biofilms, their relationship to the surface substratum or tissue, microbial physiology and species composition.
What is confocal microscopy?
Advantages of confocal microscopy for imaging biofilms and tissues
- 3D real-time imaging of live samples.
- Red, orange, green and blue lasers can target stains specific for cellular components with excitation wavelengths ranging from 458 to 633 nm.
- Detectors can be tuned to 'look for' specific signal emissions and autofluorescence between 400 an 800 nm and tune out background autofluorescence.
- Short exposure times significantly reduce photobleaching and UV killing associated with conventional epifluorescence microscopy allowing long-term continuous monitoring.
- SP2 quadruple channel detector allows monitoring of 4 stains simultaneously.
- Confocal microscopy integrates extremely well with molecular techniques such as Fluorescence In-Situ Hybridization (FISH) for studying the localization of specific types of bacteria within complex communities and Fluorescent Protein Expression (FP) for visualization of live cells without staining (the cells are genetically engineered to produce their own stain) and temporal gene expression by linking FP to the transcription of a specific gene.
Click here to read more
|
|

|
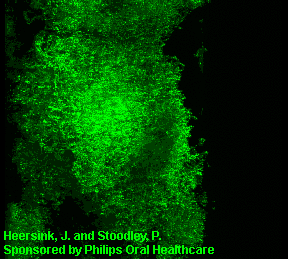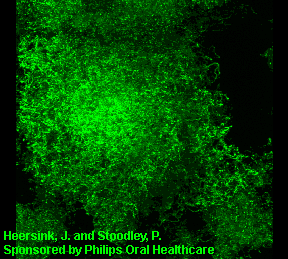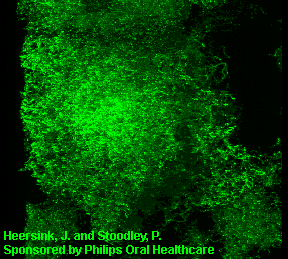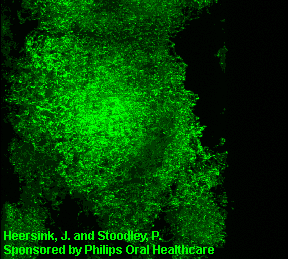
3D image of a Streptococcus mutans bacterial biofilm. S. mutans is an early colonizer of tooth surfaces and by turning the sugars and starches we eat into acid can cause caries (cavities). Staining with MolecularProbes live/dead kit stained the individual bacterial cells green indicating viability. In biofilms the bacteria often form complex cell cluster colonies consisting of 'tower', 'mushroom', and 'mound' shaped structures.
|
This image appears in Post, C.J., Stoodley, P., Hall-Stoodley, L., and Ehrlich, G.D. 2004. The role of biofilms in otolaryngologic infections. Current Opinion in Otolaryngolog & Head & Neck Surgery. 12(3):185-190.
For further reading on how this biofilm was grown refer to:
Heersink, J., Costerton, W.J. and Stoodley, P. 2003. Influence of the Sonicare® toothbrush on the structure and thickness of laboratory grown Streptococcus mutans biofilms assessed by digital time-lapse and confocal microscopy. American Journal of Dentistry. 16(2): 79-83.
Adams, H., Winston, M.T., Heersink, J., Buckingham-Meyer, K.A, Costerton, W.J., and Stoodley, P. 2002. Development of a laboratory model to assess the removal of biofilm from interproximal spaces by powered tooth brushing. American Journal of Dentistry. 15(Special issue):12B-17B.
Fluorescence Protein Technology
Expression of multiple genes and species can be monitored for involvement in biofilm formation over time using a growing family of differently colored fluorescent proteins Blue (BFP), Cyan (CFP), Green (GFP), Yellow (YFP) and Red (RFP).
GFP for tracking Quorum Sensing in Biofilms
Pseudomonas aeruginosa PAO1 genetically engineered to produce GFP when the lasB gene is expressed. LasB is a protein involved in the production of elastase, a virulence factor under control of the las-rhl QS system which is also associated with biofilm maturation. Biofilms were grown under low shear, laminar flow (left) and high shear turbulent flow (right). Under laminar flow (A) the microcolonies were mound shaped (white arrow) while under turbulent flow (B) they formed filamentous streamers (arrow). The green color indicates that QS was active in both biofilms. The construct, PAO-MH454 (lasB-gfp), was made and gifted by Morten Hentzer (DTU) and the images were taken by Laura Purevdorj-Gage in collaboration with Matt Parsek and Mary-Jo Kirisits (Northwestern).
Digital-Time Lapse Microscopy

In addition to confocal microscopy the imaging facility will house 3 epifluorescence and bright-field digital time-lapse microscopy stations. The main components of the system are the microscope, the digital camera, and computer with framestore board. Biofilms can be grown in flow cells for tracking development and behavior in real time.
What are biofilm flow cells?
|

|

What are Biofilms?

|
|

|

|
|

|

|
What are Biofilms?
Biofilms are adherent communities of bacteria surrounded by extracellular polymeric material that are found everywhere in nature, as well as in device-related and chronic human infections, such as cystic fibrosis, otitis media, endocarditis and osteomyelitis.
The ability of bacteria to attach to surfaces and organize into complex aggregates of cells reflects a fundamental survival strategy of bacteria. Bacteria within biofilms are more resistant to sub-optimal growing conditions such as poor nutrients, desiccation and the effects of antimicrobial agents and host defenses.
Studying bacteria in the environment in which they naturally grow is essential to understanding their multifaceted developmental responses including their responses to antibiotics.
|

|
|
|
Research overview:
The imaging of biofilms is fundamental to biofilm research. High-resolution confocal images provided the first evidence for the structural heterogeneity of biofilm architecture. This evidence was used to establish models of biofilm growth and ultrastructure. Imaging studies also showed that flow conditions, nutrient conditions and concentrations of signaling molecules were important in biofilm development. Such studies are crucial to a better understanding of the complex nature of biofilms, their relationship to the substratum, microbial physiology and species composition.
Technical progress in microbial genetics in the last several years has led to the development and widespread use of molecular reporters to analyze gene regulation and function. These reporter molecules function only in viable cells. Therefore analysis of fluorescent protein constructs must be consistent with the in situ analysis of viable cells.
Confocal microscopy is an essential component of imaging bacteria in biofilms because other high-resolution microscopic techniques require fixation that disrupts the three dimensional structure and kills the bacteria being analyzed. Therefore reporter gene constructs cannot be viewed after fixation. Furthermore, both conventional fluorescent microscopy and electron microscopy preclude the analysis of thick specimens such as biofilms because they fail to resolve thick samples.

Cross section made up from a composite of individual confocal images of a Streptococcus mutans biofilm grown on a glass slide. The biofilm was too thick to visualize the interior, which has been subsequently false colored in yellow. The cells on the outer edge of the biofilm were stained with MolecularProbes live/dead™ kit.
Integration of Confocal Microscopy with CGS Research

The synergy from high resolution 4D microscopy, molecular techniques, collaborations with surgeons and Carnegie Mellon MEMS engineers promises significant advancements in our understanding and treatment of interactions between the biofilm, the host and medical devices and the role of biofilms in infectious disease.
|